
The role of host proteins in SARS-CoV-2 replication
Brief description
The infection of SARS-CoV-2 causes a storm into the homeostasis of host cells due to the hijacking of a plethora of host factors required to complete the virus lifecycle. Several host proteins have been found to be involved in Protein-Protein interactions with the viral Replication Transcription Complex (RTC) composed by three viral non-structural proteins (nsps): nsp12 (RNA-dependent-RNA-polymerase), that mediates the nucleotide polymerization, and nsp8 and nsp7, acting as primase complex. In particular, findings based on proteomic studies and computational models suggested that RNA-binding proteins such as La-Related Proteins (LaRPs) and PolyA-binding proteins (PABPs) could be involved in the viral genome replication.
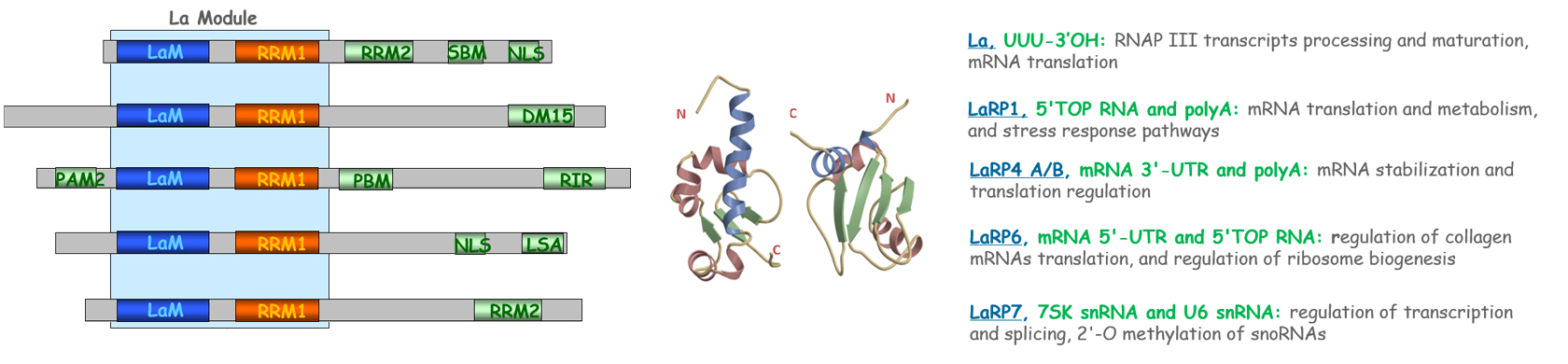
The members of the LaRPs superfamily (Genuine La, LaRP1, LaRP4A, LaRP4B, LaRP6, and LaRP7), all characterized by an extremely conserved N-terminal motif (La Module), recognize different substrates and are engaged in processes related to the cellular RNAs regulation. PABPs (PABPN1, PABPC1, PABPC3, PABPC4, PABPC5) are instead involved in mRNA length regulation and translation.
Our studies are focused to the biophysical and structural characterization of possible interactions that involve the pivot triad nsp12-8-7 of the RTC with host factors by using structural and biophysical techniques, such as Bio-Layer Interferometry (BLI), Isothermal Titration Calorimetry (ITC), Nuclear Magnetic Resonance (NMR), and cryo-Electron Microscopy (cryo-EM). We started our investigation from LaRPs to determine if one or more members of this peculiar superfamily is crucial for viral replication. By providing kinetic and structural information, our research aims to contribute for developing new therapeutic approaches.
Impact
Despite on May 2023 the WHO declared the end of global health emergency, the virus still circulates allowing the emerging of new variants and, potentially, surges in case and death. Moreover, the latest pandemic has underlined the importance to deepen the understanding of the molecular mechanism by which viruses abduct and exploit host factors. These efforts are, indeed, crucial to develop effective strategies against present and/or future aetiological agents, aiming at the prevention or mitigation of their effects on human health. In this context, a deepen knowledge of the role that host proteins play during the viral replication will contribute to figure out the molecular bases behind the infection of coronaviruses
Pipeline
-
CLINICAL
NEED -
DISEASES
ANALYSIS - DISCOVERY
-
PRECLINICAL
VALIDATION -
PRECLINICAL
DEVELOPMENT -
CLINICAL
STUDIES
Principal Investigator
Contact
Therapeutic Areas:
Product:
Drugs – Biologics
Collaborations:
- Randall Division of Cell and Molecular Biophysics, King’s College London (KCL), Regno Unito
- Institute of Biochemistry I, University of Regensburg, Germania
Scarica il pdf del progetto
